Researchers explain the imaging mechanisms of atomic force microscopy in 3D
12/07/2024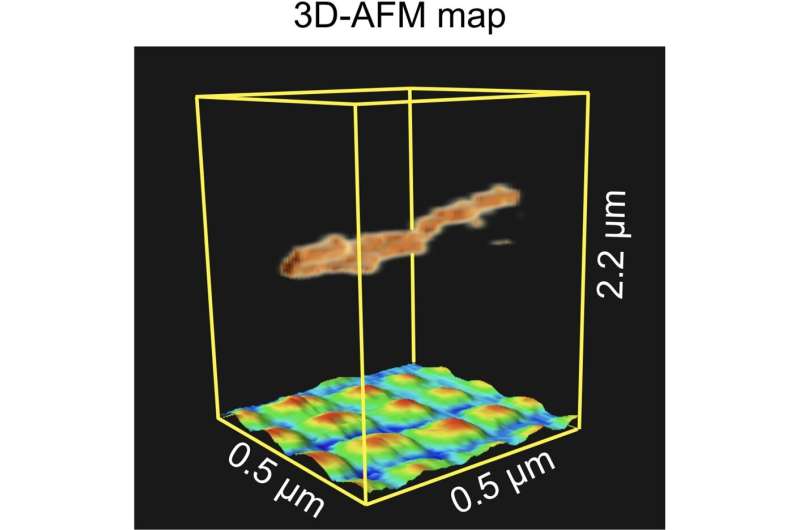
Three-dimensional atomic force microscopy (3D-AFM) map underlining its capability to image suspended flexible samples above a regular pattern of nano-sized dots. Credit: Small Methods (2024). DOI: 10.1002/smtd.202400287
Researchers at Nano Life Science Institute (WPI-NanoLSI), Kanazawa University report the 3D imaging of a suspended nanostructure. The technique used is an extension of atomic force microscopy and is a promising approach for visualizing various 3D biological systems.
Atomic force microscopy (AFM) was originally invented for visualizing surfaces with nanoscale resolution. Its basic working principle is to move an ultrathin tip over a sample's surface. During this xy-scanning motion, the tip's position in the direction perpendicular to the xy-plane follows the sample's height profile, resulting in a height map of the surface.
In recent years, ways to extend the method to 3D imaging have been explored, with researchers from Nano Life Science Institute (WPI-NanoLSI), Kanazawa University reporting pioneering experiments on living cells. However, for 3D-AFM to evolve into a widely applicable technique for visualizing flexible molecular structures, a thorough understanding of the imaging mechanisms at play is necessary.
Now, Takeshi Fukuma from Kanazawa University and colleagues have performed a detailed study of a specially designed flexible sample, providing essential insights into the theoretical basis and the interpretation of 3D-AFM experiments. The study is published in the journal Small Methods.
Using microfabrication tools, the scientists created a sample consisting of a carbon nanotube fiber resting on platinum pillars that in turn were positioned on a silicon substrate. A carbon nanotube is a structure that one can think of as a rolled-up, one-atom-thick carbon sheet. The freestanding portion of the nanotube was about 2 micrometers long. The whole structure was immersed in water, as many 3D biomolecular systems of interest occur in liquid environments.
Fukuma and colleagues then performed 3D-AFM experiments in two different modes. In static mode, the nanotip is lowered vertically towards the sample. When the tip makes contact with the suspended nanotube fiber, the latter gets pushed aside, and bends while the probe descends further. In dynamic mode, the tip, which is attached to a cantilever, is made to oscillate at a resonance frequency while being lowered.
By analyzing how the force experienced by the tip changes as a function of the tip's depth, the researchers concluded that the friction between the tip and the fiber is much larger in static mode compared to dynamic mode. The latter is therefore the mode of choice, as less friction means that potential damage to the sample is less likely.
The scientists performed computer simulations to model what happens when the tip reaches the carbon nanotube fiber. The simulations confirmed that the suspended nanotube displaces laterally, and that a continuously vibrating tip (as in dynamical mode) results in weaker forces experienced by the sample, hindering strong adhesion of the tip to the fiber.
Fukuma and colleagues then performed experiments with a carbon nanotube fiber suspended above a regular pattern of nano-sized platinum dots deposited on a silicon substrate. The measurements were done in dynamical mode. The reconstructed 3D map of the scanned volume clearly showed the fiber and the dots below it, underlining the capability of 3D-AFM to image vertically overlapping nanostructures.
These findings show that AFM can generally be applied to visualize flexible 3D structures. "The advancements made in this study may potentially lead to more detailed and accurate AFM analysis of various 3D biological systems such as cells, organelles, chromosomes, and vesicles," state the scientists.
Source: https://tinyurl.com/3xt6c2xs via Phys.Org

